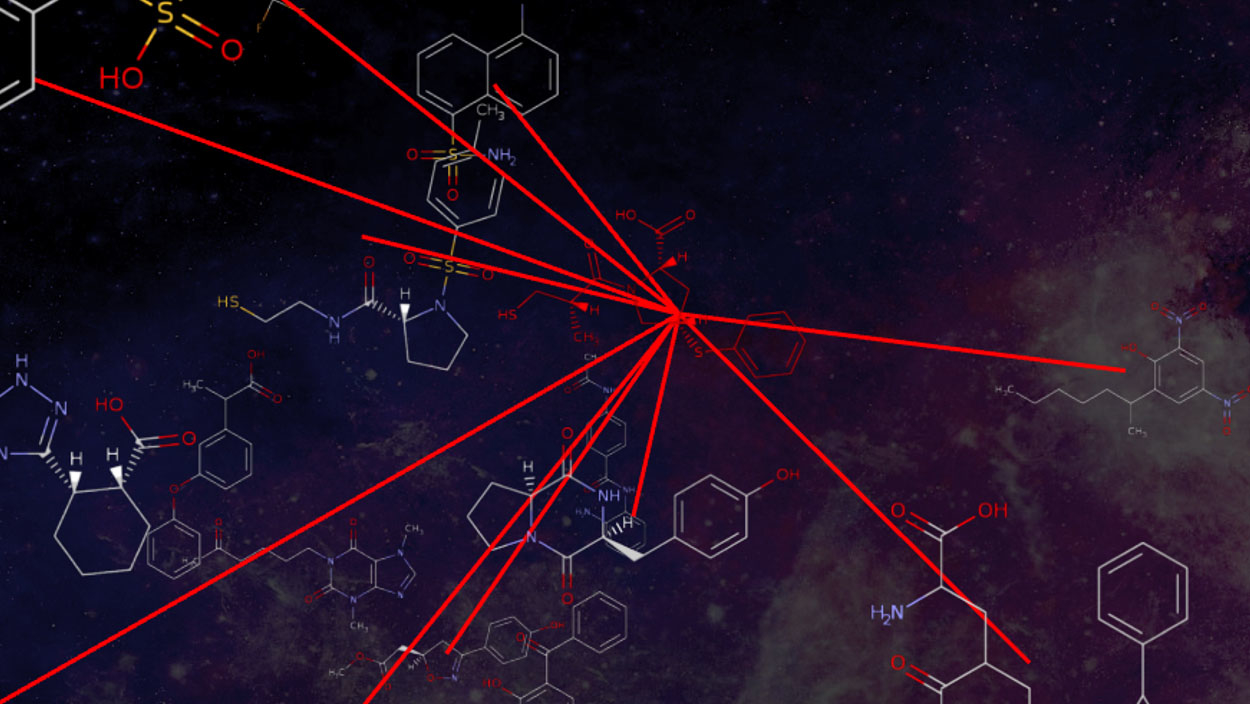
Researchers from North Carolina State University have created a new online service – ChemMaps – that allows users to interactively navigate the chemical space of over 8,000 drugs and 47,000 environmental compounds in 3D and real time. ChemMaps is designed to be a central resource for students and researchers who want to easily visualize and study complicated sets of chemical structures. The first release of the free-to-use website is available at www.chemmaps.com.
“The premise for ChemMaps was to make it the Google Maps of drugs,” says Denis Fourches, assistant professor of chemistry at NC State. “It allows anyone using a computer or a tablet to easily browse and navigate the chemical space of marketed drugs in a 3D environment. We really wanted to make the graphic interface in ChemMaps relevant for a student or for a specialist in medicinal chemistry.”
With ChemMaps, the chemicals look like stars – points of light scattered across the screen. Each star, or compound, is positioned in relationship to the others within the complex chemical space based on their structural properties. When a user clicks on a particular compound-star, several key characteristics of that chemical are displayed: its systematic name, brand name, chemical structure, medical indication, external identifiers, and other physical chemical properties.
Currently, there are two main maps available. The DrugMap includes around 8,000 drugs (marketed, in development and withdrawn) and was prepared from the DrugBank database. The environmental map, or EnvMap, includes over 47,000 compounds (such as pesticides or flame retardants) of relevance for the National Institute of Environmental Health Studies (NIEHS) and the United States Environmental Protection Agency (EPA) and was prepared from the Tox21, Toxcast, and other NIEHS databases.
The researchers plan to add features over time, including the ability for collaborators and companies to visualize and project proprietary sets of chemicals onto the ChemMaps. They also plan to make the maps even more interactive, easy to manipulate, and compatible with augmented and virtual reality devices.
“This is the first tool in the public domain that allows anyone, particularly non-experts in cheminformatics, to easily, visually and quickly access all this complex chemistry information in one place,” Fourches says. “This tool could help researchers better understand the chemical neighborhood of a particular compound. In fact, this is critical for drug repurposing or chemical risk assessment.”
The research appears in Bioinformatics. Dr. Alex Borrel, former NC State postdoctoral fellow currently at the NIEHS and first author of the paper, created the program. Fourches conceived ChemMaps and is corresponding author. Dr. Nicole Kleinstreuer, computational biologist at the NIEHS, also contributed to the work.
-peake-
Note to editors: An abstract of the paper follows
“Exploring Drug Space with ChemMaps.com”
DOI: 10.1093/bioinformatics/bty412
Authors: Alexandre Borrel and Denis Fourches, North Carolina State University; Alexandre Borrel and Nicole Kleinstreuer, National Institute of Environmental Health Sciences
Published: Online in Bioinformatics
Abstract:
Easily navigating chemical space has become more important due to the increasing size and diversity of publicly-accessible databases such as DrugBank, ChEMBL, or Tox21. To do so, modelers typically rely on complex projection techniques using molecular descriptors computed for all the chemicals to be visualized. However, the multiple cheminformatics steps required to prepare, characterize, compute and explore those molecules, are technical, typically necessitate scripting skills, and thus represent a real obstacle for non-specialists. We developed the ChemMaps.com webserver to easily browse, navigate, and mine chemical space. The first version of ChemMaps.com features more than 8,000 approved, in development, and rejected drugs, as well as over 47,000 environmental chemicals. The webserver is freely available at https://www.chemmaps.com