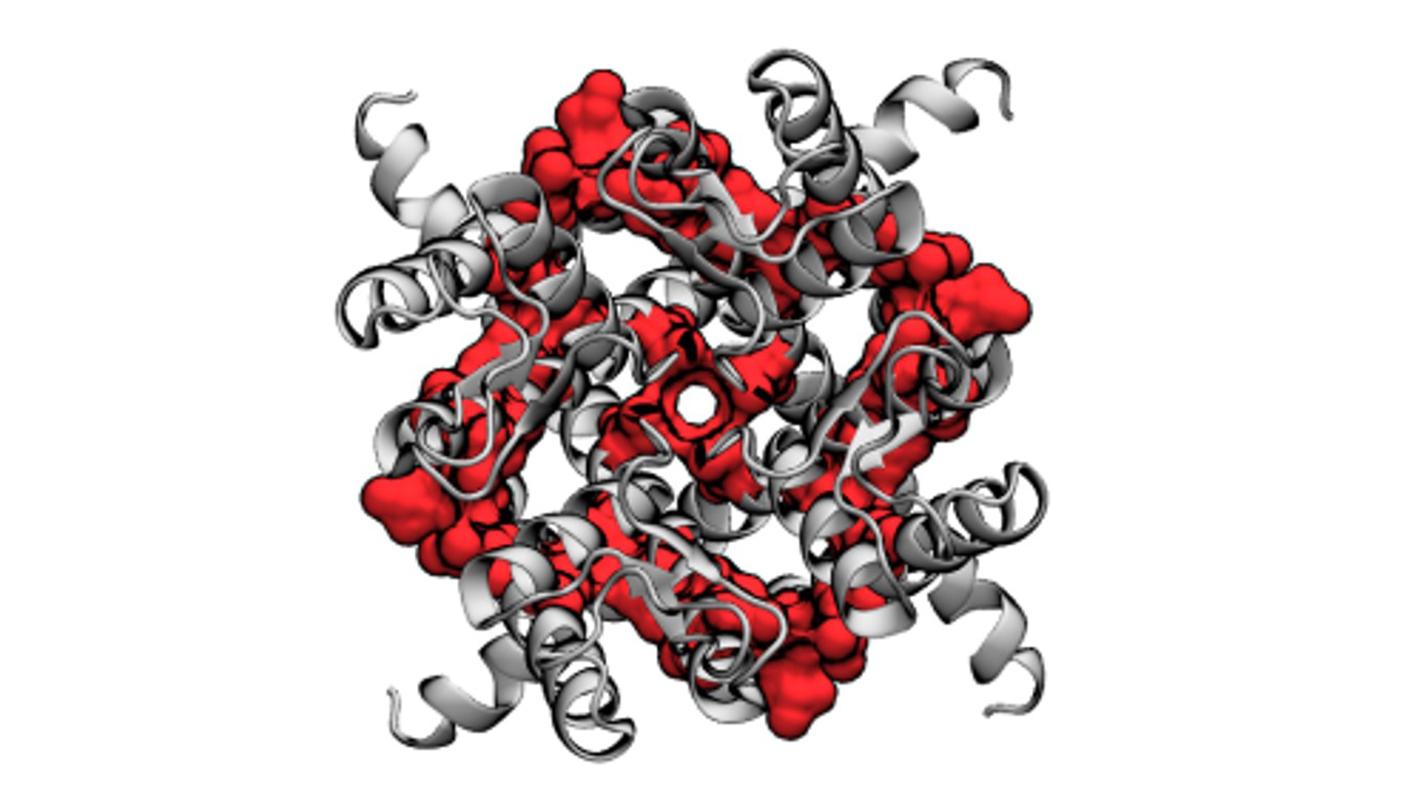
Using a computer model, researchers from North Carolina State University and the University of Illinois at Chicago have revealed the effect of increased amounts of cholesterol on a specific ion channel involved in regulating potassium levels in the heart. The work sheds further light on interactions between cholesterol and heart function and could have an impact on future cardiac therapies.
Ion channels are proteins located within a cell membrane that control the transport of ions between a cell’s surrounding environment and the interior of the cell. The electrical current that allows the heart muscle to contract is a product of a series of ion transfers across the cell membrane. Every cardiac cell has ion channels in the membrane that transport a specific charged atom – such as calcium, sodium, or potassium – from the external environment into the heart cell.
Belinda Akpa, assistant professor of integrated synthetic and systems biology and electrical and computer engineering at NC State and co-corresponding author of a paper describing the research, looked at the effect of cholesterol molecules on a particular ion channel, called Kir2, which regulates the transfer of potassium into cardiac cells.
“Cholesterol isn’t in and of itself a bad thing,” Akpa says. “It’s always present in the cell membrane. When the levels of cholesterol change we start to have problems. Given that cholesterol is something like 30 percent of a normal membrane, we wanted to understand why a relatively small increase – going from about 30 to 40 percent – suddenly makes things go wrong.”
Akpa, University of Chicago at Illinois Ph.D. student Nicolas Barbera, and co-corresponding author Irena Levitan, professor of medicine, pharmacology, and bioengineering at the University of Illinois at Chicago, used computer modeling to reveal the ways in which cholesterol molecules interact with the Kir2 ion channel. They found that while individual cholesterol molecules don’t bind strongly to the Kir2 channel, increasing the levels of cholesterol made these interactions more numerous, essentially overwhelming the channel.
Proteins and small molecules often interact like locks and keys, where only a specific molecule can fit into a particular region on the protein. These interactions cause the protein to change shape – in the case of Kir2, cholesterol sliding into these regions interferes with the protein’s attempts to open or close to allow potassium ions into a cardiac cell. In their model, Akpa, Barbera and Levitan identified four “locks” on the Kir2 ion channel that cholesterol molecules attempted to occupy.
“Cholesterol may actually belong in some of these locks, but we’re also seeing it try to move into places that should probably be unoccupied, which interferes with the ability of the protein to change its shape in a way that allows it to open and close normally,” says Akpa. “This is a problem because cells require ion channels to orchestrate a sophisticated choreography of ions moving in and out at different times. Essentially, it’s like taking a symphony of ion exchanges and inserting a wrong note.”
Future work for the team will focus specifically on how additional cholesterol changes the ability of the protein to open and close.
The work appears in Biophysical Journal, and was funded by the National Institutes of Health (grants HL-073965, HL-083298, and T32 HL-82547. Barbera is first author. Manuela Ayee from the University of Illinois at Chicago also contributed to the work.
-peake-
Note to editors: An abstract of the paper follows.
“Molecular Dynamics Simulations of Kir2.2 Interactions with an Ensemble of Cholesterol Molecules”
DOI: 10.1016/j.bpj.2018.07.041
Authors: Belinda Akpa, North Carolina State University; Nicolas Barbera, Manuela Ayee, Irena Levitan, University of Illinois at Chicago
Published: Biophysical Journal
Abstract:
Cholesterol is a major regulator of multiple types of ion channels, but the specific mechanisms and the dynamics of its interactions with the channels are not well understood. Kir2 channels were shown to be sensitive to cholesterol through direct interactions with “cholesterol-sensitive” regions on the channel protein. In this work, we used MARTINI coarse-grained simulations to analyze the long (µs) timescale dynamics of cholesterol with Kir2.2 channels embedded into a model membrane containing POPC phospholipid with 30 mol% cholesterol. This approach allows us to simulate the dynamic, unbiased migration of cholesterol molecules from the lipid membrane environment to the protein surface of Kir2.2 and explore the favorability of cholesterol interactions at both surface sites and recessed pockets of the channel. We found that the cholesterol environment surrounding Kir channels forms a complex milieu of different short- and longterm interactions, with multiple cholesterol molecules concurrently interacting with the channel. Furthermore, utilizing principles from network theory, we identified four discrete cholesterol binding sites within the previously identified cholesterol sensitive region, which exist depending on the conformational state of the channel – open or closed. We also discovered that a 2-fold decrease in the cholesterol level of the membrane, which we found earlier to increase Kir2 activity, results in a site-specific decrease of cholesterol occupancy at these sites in both the open and closed states: cholesterol molecules at the deepest of these discrete sites shows no change in occupancy at different cholesterol levels, while the remaining sites showed a marked decrease in occupancy.